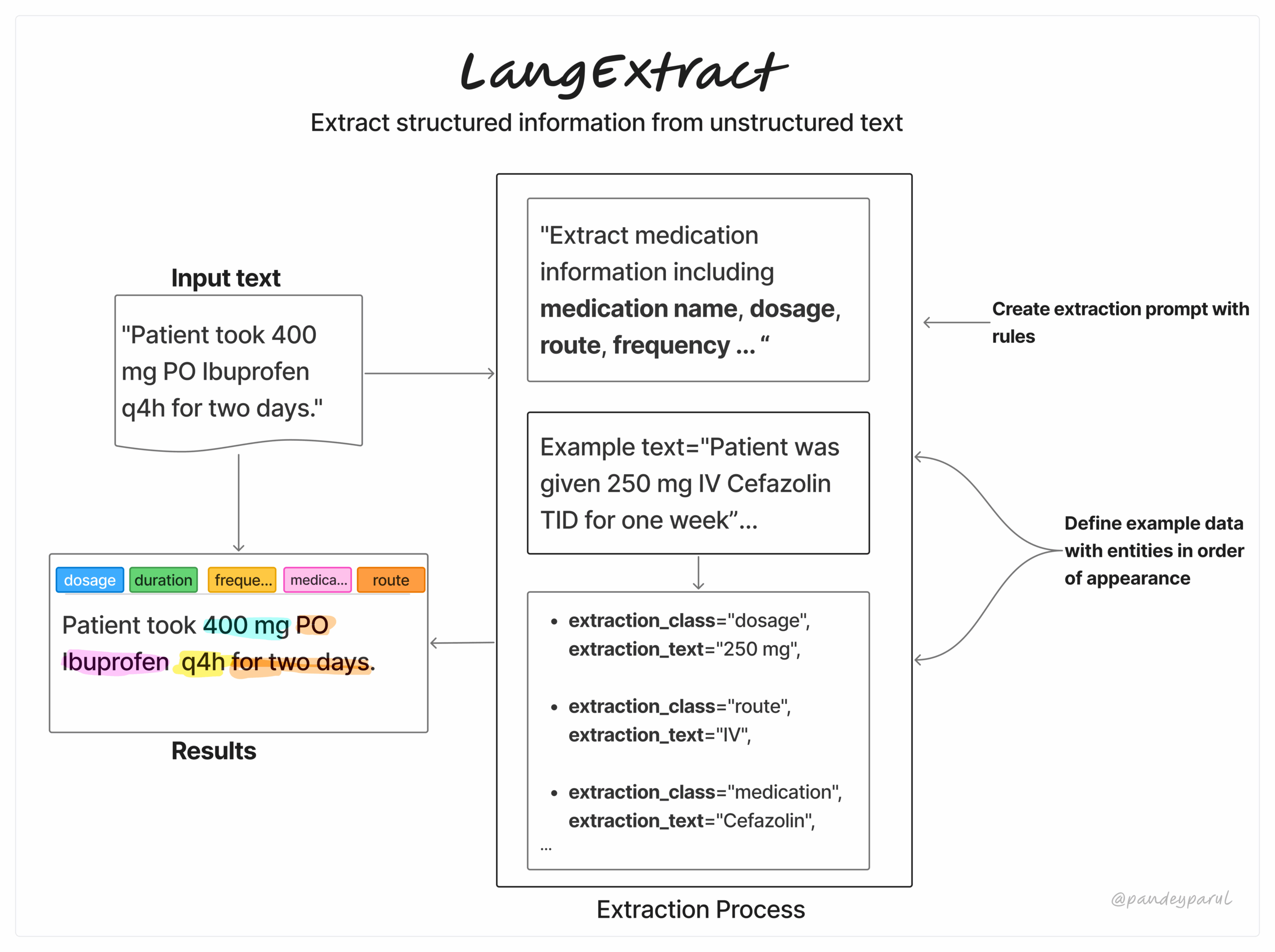
LangExtract is a from developers at Google that makes it easy to turn messy, unstructured text into clean, structured data by leveraging LLMs. Users can provide a few few-shot examples along with a custom schema and get results based on that. It works both with proprietary as well as local LLMs (via Ollama).
A significant amount of data in healthcare is unstructured, making it an ideal area where a tool like this can be beneficial. Clinical notes are long and full of abbreviations and inconsistencies. Important details such as drug names, dosages, and especially adverse drug reactions (ADRs) get buried in the text. Therefore, for this article, I wanted to see if LangExtract could handle adverse drug reaction (ADR) detection in clinical notes. More importantly, is it effective? Let’s find out in this article. Note that while LangExtract is an open-source project from developers at Google, it is not an officially supported Google product.
Just a quick note: I’m only showing how LangExtract works. I’m not a doctor, and this isn’t medical advice.
▶️ Here is a detailed Kaggle notebook to follow along.
Why ADR Extraction Matters
An Adverse Drug Reaction (ADR) is a harmful, unintended result caused by taking a medication. These can range from mild side effects like nausea or dizziness to severe outcomes that may require medical attention.

Detecting them quickly is critical for patient safety and pharmacovigilance. The challenge is that in clinical notes, ADRs are buried alongside past conditions, lab results, and other context. As a result, detecting them is tricky. Using LLMs to detect ADRs is an ongoing area of research. Some recent works have shown that LLMs are good at raising red flags but not reliable. So, ADR extraction is a good stress test for LangExtract, as the goal here is to see if this library can spot the adverse reactions amongst other entities in clinical notes like medications, dosages, severity, etc.
How LangExtract Works
Before we jump into usage, let’s break down LangExtract’s workflow. It’s a simple three-step process:
- Define your extraction task by writing a clear prompt that specifies exactly what you want to extract.
- Provide a few high-quality examples to guide the model towards the format and level of detail you expect.
- Submit your input text, choose the model, and let LangExtract process it. Users can then review the results, visualize them, or pass them directly into their downstream pipeline.
The official GitHub repository of the tool has detailed examples spanning multiple domains, from entity extraction in Shakespeare’s Romeo & Juliet to medication identification in clinical notes and structuring radiology reports. Do check them out.
Installation
First we need to install the LangExtract library. It’s always a good idea to do this within a virtual environment to keep your project dependencies isolated.
pip install langextractIdentifying Adverse Drug Reactions in Clinical Notes with LangExtract & Gemini
Now let’s get to our use case. For this walkthrough, I’ll use Google’s Gemini 2.5 Flash model. You could also use Gemini Pro for more complex reasoning tasks. You’ll need to first set your API key:
export LANGEXTRACT_API_KEY="your-api-key-here"▶️ Here is a detailed Kaggle notebook to follow along.
Step 1: Define the Extraction Task
Let’s create our prompt for extracting medications, dosages, adverse reactions, and actions taken. We can also ask for severity where mentioned.
prompt = textwrap.dedent("""\
Extract medication, dosage, adverse reaction, and action taken from the text.
For each adverse reaction, include its severity as an attribute if mentioned.
Use exact text spans from the original text. Do not paraphrase.
Return entities in the order they appear.""")
Next, let’s provide an example to guide the model towards the correct format:
# 1) Define the prompt
prompt = textwrap.dedent("""\
Extract condition, medication, dosage, adverse reaction, and action taken from the text.
For each adverse reaction, include its severity as an attribute if mentioned.
Use exact text spans from the original text. Do not paraphrase.
Return entities in the order they appear.""")
# 2) Example
examples = [
lx.data.ExampleData(
text=(
"After taking ibuprofen 400 mg for a headache, "
"the patient developed mild stomach pain. "
"They stopped taking the medicine."
),
extractions=[
lx.data.Extraction(
extraction_class="condition",
extraction_text="headache"
),
lx.data.Extraction(
extraction_class="medication",
extraction_text="ibuprofen"
),
lx.data.Extraction(
extraction_class="dosage",
extraction_text="400 mg"
),
lx.data.Extraction(
extraction_class="adverse_reaction",
extraction_text="mild stomach pain",
attributes={"severity": "mild"}
),
lx.data.Extraction(
extraction_class="action_taken",
extraction_text="They stopped taking the medicine"
)
]
)
]Step 2: Provide the Input and Run the Extraction
For the input, I’m using a real clinical sentence from the ADE Corpus v2 dataset on Hugging Face.
input_text = (
"A 27-year-old man who had a history of bronchial asthma, "
"eosinophilic enteritis, and eosinophilic pneumonia presented with "
"fever, skin eruptions, cervical lymphadenopathy, hepatosplenomegaly, "
"atypical lymphocytosis, and eosinophilia two weeks after receiving "
"trimethoprim (TMP)-sulfamethoxazole (SMX) treatment."
)Next, let’s run LangExtract with the Gemini-2.5-Flash model.
result = lx.extract(
text_or_documents=input_text,
prompt_description=prompt,
examples=examples,
model_id="gemini-2.5-flash",
api_key=LANGEXTRACT_API_KEY
)Step 3: View the Results
You can display the extracted entities with positions
print(f"Input: {input_text}\n")
print("Extracted entities:")
for entity in result.extractions:
position_info = ""
if entity.char_interval:
start, end = entity.char_interval.start_pos, entity.char_interval.end_pos
position_info = f" (pos: {start}-{end})"
print(f"• {entity.extraction_class.capitalize()}: {entity.extraction_text}{position_info}")
LangExtract correctly identifies the adverse drug reaction without confusing it with the patient’s pre-existing conditions, which is a key challenge in this type of task.
If you want to visualize it, it’s going to create this .jsonl file. You can load that .jsonl file by calling the visualization function, and it will create an HTML file for you.
lx.io.save_annotated_documents(
[result],
output_name="adr_extraction.jsonl",
output_dir="."
)
html_content = lx.visualize("adr_extraction.jsonl")
# Display the HTML content directly
display((html_content))
Working with longer clinical notes
Real clinical notes are often much longer than the example shown above. For instance, here is an actual note from the ADE-Corpus-V2 dataset released under the MIT License. You can access it on Hugging Face or Zenodo.

To process longer texts with LangExtract, you keep the same workflow but add three parameters:
extraction_passes runs multiple passes over the text to catch more details and improve recall.
max_workers controls parallel processing so larger documents can be handled faster.
max_char_buffer splits the text into smaller chunks, which helps the model stay accurate even when the input is very long.
result = lx.extract(
text_or_documents=input_text,
prompt_description=prompt,
examples=examples,
model_id="gemini-2.5-flash",
extraction_passes=3,
max_workers=20,
max_char_buffer=1000
)Here is the output. For brevity, I’m only showing a portion of the output here.

If you want, you can also pass a document’s URL directly to the text_or_documents parameter.
Using LangExtract with Local models via Ollama
LangExtract isn’t limited to proprietary APIs. You can also run it with local models through Ollama. This is especially useful when working with privacy-sensitive clinical data that can’t leave your secure environment. You can set up Ollama locally, pull your preferred model, and point LangExtract to it. Full instructions are available in the official docs.
Conclusion
If you’re building an information retrieval system or any application involving metadata extraction, LangExtract can save you a significant amount of preprocessing effort. In my ADR experiments, LangExtract performed well, correctly identifying medications, dosages, and reactions. What I noticed is that the output directly depends on the quality of the few-shot examples provided by the user, which means while LLMs do the heavy lifting, humans still remain an important part of the loop. The results were encouraging, but since clinical data is high-risk, broader and more rigorous testing across diverse datasets is still needed before moving toward production use.
Source link
#LangExtract #Turn #Messy #Clinical #Notes #Structured #Data